-Search query
-Search result
Showing 1 - 50 of 128 items for (author: chio & us)

EMDB-42977: 
Cryo-EM structure of singly-bound SNF2h-nucleosome complex with SNF2h at inactive SHL2 (conformation 1)
Method: single particle / : Chio US, Palovcak E, Armache JP, Narlikar GJ, Cheng Y

EMDB-43000: 
Cryo-EM structure of SNF2h-nucleosome complex (consensus structure)
Method: single particle / : Chio US, Palovcak E, Armache JP, Narlikar GJ, Cheng Y

EMDB-43001: 
Cryo-EM structure of SNF2h-nucleosome complex (single-bound structure)
Method: single particle / : Chio US, Palovcak E, Armache JP, Narlikar GJ, Cheng Y

EMDB-43002: 
Cryo-EM structure of doubly-bound SNF2h-nucleosome complex
Method: single particle / : Chio US, Palovcak E, Armache JP, Narlikar GJ, Cheng Y

EMDB-43003: 
Cryo-EM structure of singly-bound SNF2h-nucleosome complex with SNF2h at inactive SHL2 (conformation 2)
Method: single particle / : Chio US, Palovcak E, Armache JP, Narlikar GJ, Cheng Y

EMDB-43004: 
Cryo-EM structure of doubly-bound SNF2h-nucleosome complex (conformation 1)
Method: single particle / : Chio US, Palovcak E, Armache JP, Narlikar GJ, Cheng Y

EMDB-43005: 
Cryo-EM structure of doubly-bound SNF2h-nucleosome complex (conformation 2)
Method: single particle / : Chio US, Palovcak E, Armache JP, Narlikar GJ, Cheng Y

EMDB-18179: 
Human KMN network (outer kinetochore)
Method: single particle / : Raisch T, Polley S, Vetter I, Musacchio A, Raunser S

PDB-8q5h: 
Human KMN network (outer kinetochore)
Method: single particle / : Raisch T, Polley S, Vetter I, Musacchio A, Raunser S

EMDB-17241: 
C. elegans L1 80S ribosome
Method: subtomogram averaging / : Schioetz OH, Kaiser CJO, Klumpe S, Beck F, Plitzko JM

EMDB-17242: 
C. elegans L1 larva 80S ribosome class 1
Method: subtomogram averaging / : Schioetz OH, Kaiser CJO, Klumpe S, Beck F, Plitzko JM

EMDB-17243: 
C. elegans L1 larva 80S ribosome class 2
Method: subtomogram averaging / : Schioetz OH, Kaiser CJO, Klumpe S, Beck F, Plitzko JM

EMDB-17244: 
C. elegans L1 larva 80S ribosome class 3
Method: subtomogram averaging / : Schioetz OH, Kaiser CJO, Klumpe S, Beck F, Plitzko JM

EMDB-17245: 
C. elegans L1 larva 80S ribosome class 4
Method: subtomogram averaging / : Schioetz OH, Kaiser CJO, Klumpe S, Beck F, Plitzko JM

EMDB-17246: 
C. elegans L1 larva body wall muscle tomogram
Method: electron tomography / : Schioetz OH, Kaiser CJO, Klumpe S, Beck F, Plitzko JM

EMDB-17247: 
C. elegans L1 larva ventral pharyngeal periphery tomogram
Method: electron tomography / : Schioetz OH, Kaiser CJO, Klumpe S, Beck F, Plitzko JM

EMDB-17248: 
Tomogram of the nucleus of a C. elegans L1 larva
Method: electron tomography / : Schioetz OH, Kaiser CJO, Klumpe S, Beck F, Plitzko JM

EMDB-18186: 
C. elegans L1 larva
Method: electron tomography / : Schioetz OH, Kaiser CJO, Klumpe S, Klebl DP, Schneider J, Beck F, Plitzko JM

EMDB-18187: 
Focused refinment of 11-protofilament microtubule from C. elegans
Method: subtomogram averaging / : Schioetz OH, Kaiser CJO, Klumpe S, Klebl DP, Schneider J, Beck F, Plitzko JM

EMDB-17187: 
48-nm repeat of the native axonemal doublet microtubule from bovine sperm
Method: single particle / : Leung MR, Zeng J, Zhang R, Zeev-Ben-Mordehai T

PDB-8otz: 
48-nm repeat of the native axonemal doublet microtubule from bovine sperm
Method: single particle / : Leung MR, Zeng J, Zhang R, Zeev-Ben-Mordehai T

EMDB-15475: 
Structure of the Ancestral Scaffold Antigen-5 of Coronavirus Spike protein
Method: single particle / : Hueting D, Schriever K, Wallden K, Andrell J, Syren PO

EMDB-15482: 
Structure of the Ancestral Scaffold Antigen-6 of Coronavirus Spike protein
Method: single particle / : Hueting D, Schriever K, Wallden K, Andrell J, Syren PO

PDB-8aja: 
Structure of the Ancestral Scaffold Antigen-5 of Coronavirus Spike protein
Method: single particle / : Hueting D, Schriever K, Wallden K, Andrell J, Syren PO

PDB-8ajl: 
Structure of the Ancestral Scaffold Antigen-6 of Coronavirus Spike protein
Method: single particle / : Hueting D, Schriever K, Wallden K, Andrell J, Syren PO
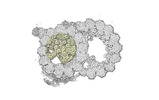
EMDB-18071: 
48-nm repeat of the native axonemal doublet microtubule from bovine sperm, local refinement A-lumen top
Method: single particle / : Leung MR, Zeng J, Zhang R, Zeev-Ben-Mordehai T

EMDB-18072: 
48-nm repeat of the native axonemal doublet microtubule from bovine sperm, local refinement A-lumen middle
Method: single particle / : Leung MR, Zeng J, Zhang R, Zeev-Ben-Mordehai T

EMDB-18073: 
48-nm repeat of the native axonemal doublet microtubule from bovine sperm, local refinement A-lumen bottom
Method: single particle / : Leung MR, Zeng J, Zhang R, Zeev-Ben-Mordehai T

EMDB-18074: 
48-nm repeat of the native axonemal doublet microtubule from bovine sperm, local refinement A01-A02 top
Method: single particle / : Leung MR, Zeng J, Zhang R, Zeev-Ben-Mordehai T
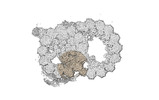
EMDB-18075: 
48-nm repeat of the native axonemal doublet microtubule from bovine sperm, local refinement A01-A02 middle
Method: single particle / : Leung MR, Zeng J, Zhang R, Zeev-Ben-Mordehai T

EMDB-18076: 
48-nm repeat of the native axonemal doublet microtubule from bovine sperm, local refinement A01-A02 bottom
Method: single particle / : Leung MR, Zeng J, Zhang R, Zeev-Ben-Mordehai T
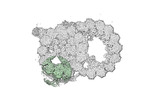
EMDB-18077: 
48-nm repeat of the native axonemal doublet microtubule from bovine sperm, local refinement A02-A04 top
Method: single particle / : Leung MR, Zeng J, Zhang R, Zeev-Ben-Mordehai T
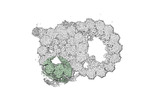
EMDB-18078: 
48-nm repeat of the native axonemal doublet microtubule from bovine sperm, local refinement A02-A04 middle
Method: single particle / : Leung MR, Zeng J, Zhang R, Zeev-Ben-Mordehai T

EMDB-18079: 
48-nm repeat of the native axonemal doublet microtubule from bovine sperm, local refinement A02-A04 bottom
Method: single particle / : Leung MR, Zeng J, Zhang R, Zeev-Ben-Mordehai T
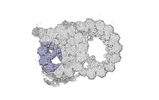
EMDB-18080: 
48-nm repeat of the native axonemal doublet microtubule from bovine sperm, local refinement A04-A06 top
Method: single particle / : Leung MR, Zeng J, Zhang R, Zeev-Ben-Mordehai T
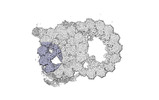
EMDB-18081: 
48-nm repeat of the native axonemal doublet microtubule from bovine sperm, local refinement A04-A06 middle
Method: single particle / : Leung MR, Zeng J, Zhang R, Zeev-Ben-Mordehai T
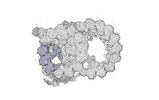
EMDB-18082: 
48-nm repeat of the native axonemal doublet microtubule from bovine sperm, local refinement A04-A06 bottom
Method: single particle / : Leung MR, Zeng J, Zhang R, Zeev-Ben-Mordehai T
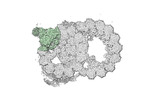
EMDB-18083: 
48-nm repeat of the native axonemal doublet microtubule from bovine sperm, local refinement A07-A08 top
Method: single particle / : Leung MR, Zeng J, Zhang R, Zeev-Ben-Mordehai T
EMDB-18084: 
48-nm repeat of the native axonemal doublet microtubule from bovine sperm, local refinement A07-A08 middle
Method: single particle / : Leung MR, Zeng J, Zhang R, Zeev-Ben-Mordehai T

EMDB-18085: 
48-nm repeat of the native axonemal doublet microtubule from bovine sperm, local refinement A07-A08 bottom
Method: single particle / : Leung MR, Zeng J, Zhang R, Zeev-Ben-Mordehai T

EMDB-18086: 
48-nm repeat of the native axonemal doublet microtubule from bovine sperm, local refinement Outer Junction top
Method: single particle / : Leung MR, Zeng J, Zhang R, Zeev-Ben-Mordehai T
EMDB-18087: 
48-nm repeat of the native axonemal doublet microtubule from bovine sperm, local refinement Outer Junction middle
Method: single particle / : Leung MR, Zeng J, Zhang R, Zeev-Ben-Mordehai T
EMDB-18088: 
48-nm repeat of the native axonemal doublet microtubule from bovine sperm, local refinement Outer Junction bottom
Method: single particle / : Leung MR, Zeng J, Zhang R, Zeev-Ben-Mordehai T
EMDB-18089: 
48-nm repeat of the native axonemal doublet microtubule from bovine sperm, local refinement Ribbon top
Method: single particle / : Leung MR, Zeng J, Zhang R, Zeev-Ben-Mordehai T
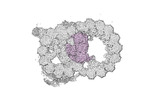
EMDB-18090: 
48-nm repeat of the native axonemal doublet microtubule from bovine sperm, local refinement Ribbon middle
Method: single particle / : Leung MR, Zeng J, Zhang R, Zeev-Ben-Mordehai T

EMDB-18091: 
48-nm repeat of the native axonemal doublet microtubule from bovine sperm, local refinement Ribbon bottom
Method: single particle / : Leung MR, Zeng J, Zhang R, Zeev-Ben-Mordehai T
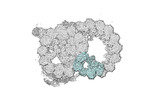
EMDB-18092: 
48-nm repeat of the native axonemal doublet microtubule from bovine sperm, local refinement Inner Junction top
Method: single particle / : Leung MR, Zeng J, Zhang R, Zeev-Ben-Mordehai T
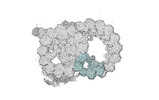
EMDB-18093: 
48-nm repeat of the native axonemal doublet microtubule from bovine sperm, local refinement Inner Junction middle
Method: single particle / : Leung MR, Zeng J, Zhang R, Zeev-Ben-Mordehai T
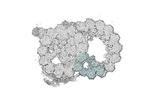
EMDB-18094: 
48-nm repeat of the native axonemal doublet microtubule from bovine sperm, local refinement Inner Junction bottom
Method: single particle / : Leung MR, Zeng J, Zhang R, Zeev-Ben-Mordehai T
EMDB-18095: 
48-nm repeat of the native axonemal doublet microtubule from bovine sperm, local refinement B01-B03 top
Method: single particle / : Leung MR, Zeng J, Zhang R, Zeev-Ben-Mordehai T
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
